Microbial evolution and systematics
Planet earth is approximately 4.6 Billions year old. Prokaryotic cell’s fossilized remains were found in sedimentary rocks and stromatolites which were estimated to be 3.5 to 3.8 Billions years old. Earliest prokaryotes were anaerobes.
Cyanobacteria and oxygen producing photosynthesis developed approximately 2.5 to 3.0 Billions years ago as the oxygen become plentiful, microbial diversity increased.
Carl Woese and colleagues studied 16S rRNA gene is approximately 1550 bases in length and prossesses region in which sequences are conserved which helps in sequence alignment and variable and hyper variable regions which helps in discrimination from one another.
Based on 16S rRNA studies Woese organized three domains which were based on evolutionary relationship namely Bacteria, Archaea and Eukarya. Woese put all eukaryotic kingdoms i.e. Protista, Fungi, Plantar, Animalia into a single domain. Woese divided the kingdom of Monera into the Bacteria and the Archaea.
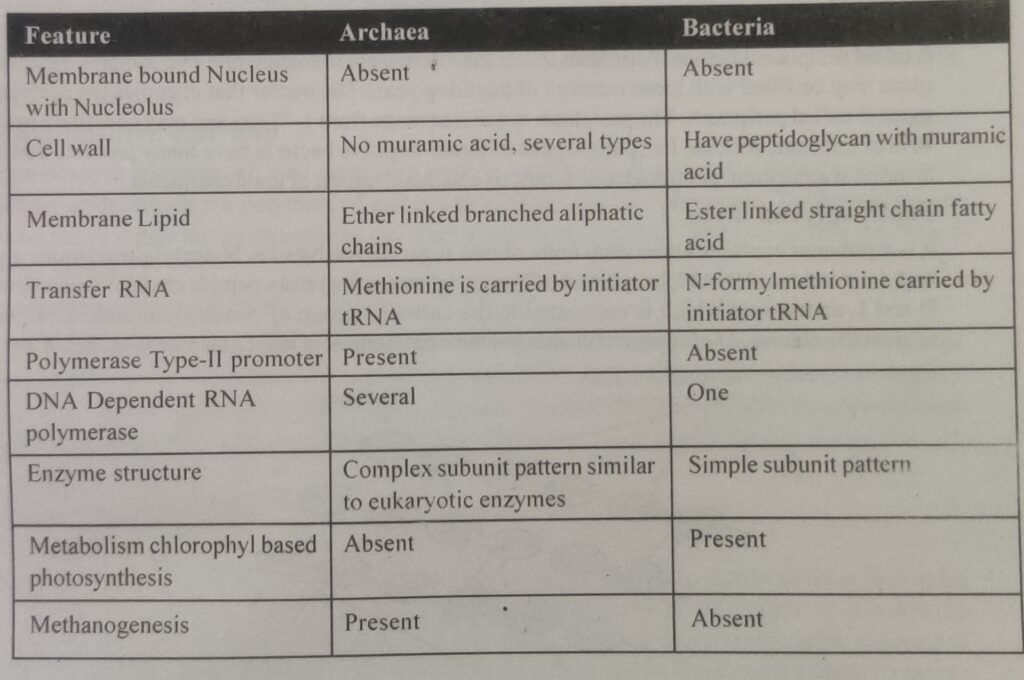
Comparison : Archaea and Bacteria
Table of Contents
BACTERIAL CELL WALL
Bacterial cell wall is rigid and present in almost all the prokaryotic cell’s except for the Mycoplasma, L-form bacteria and some Archaea.
Bacterial cell wall is made up of peptidoglycan or murein layer outside the plasma membrane Gram positive bacterial cell wall consists of 20-80 nm thick peptidoglycan layer.
Gram nagative cell wall have 2-7 nm thick peptidoglycan layer surrounded by 7-8 nm thick outer membrane.
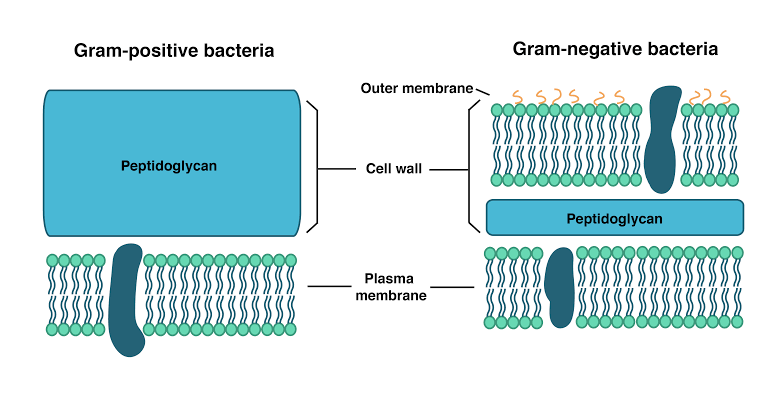
Difference between Gram positive and Gram nagative Bacterium
a space is present between the plasma membrane and outer membrane in gram-negative bacteria is called periplasmic space. Periplasm space, may be present in some gram positive Bacteria.
Periplasmic space mein may be fillied with loose network of peptidoglycan. The matter that occupies the periplasmic space is called periplasmic.
The periplasmic space of Gram nagative Bacterium have many proteins that helps in nutrient acquisition, peptidoglycan synthesis and modification of toxic chemicals.
Peptidoglycan layer
It is a polymer contains alternating units of two sugar derivatives i.e. N- acetylgluconsamine and N- acetylmuramic acid (NAM, Lactyl ether of N- acetylglucosamine) and a peptide chain of four alternating D and L- amino acid which is connected to the carboxyl group of N- acetylmuramic acid. Amino acids are L- alanine, D- glutamic acid, meso- diaminopimelic acid, meso-diaminopimelic acid and D- alanine.
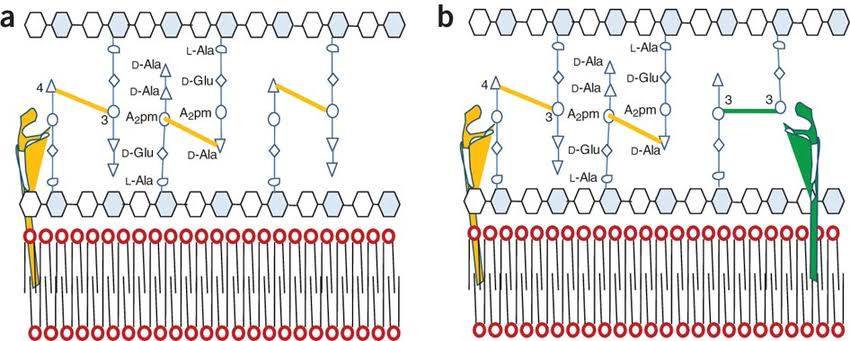
D- glutamic acid, meso-diaminopimelic acid (DAP) and D- alanine are not present in protein. Presence of D- amino acids in cell wall of Bacteria make them resistant to the action of proteolytic enzymes. In some Bacteria L- lysine is present as third resistant instead of DAP.
These peptidoglycan subunit are joined by cross links between the peptide mostly the carboxyl I group of the termainal D alanine is connected directly to the amino group of diaminopimelic acid (DAP) but sometimes peptide interbridge is also present. Peptide interbridge is mostly absent in Gram nagative Bacteria.
Cross linking provides shape and integrity to the Gram positive Bacteria cell and the sometime make then elastic and stretchable.
Gram positive cell wall
Gram positive Bacteria cell wall contains a peptide inter bridge. It also contains large amounts of teichoic acids, vichar polymer of glycerol or ribitol joined by phosphate. Teichoic acids are r connect to the the peptidoglycan aur plasma membrane lipids.
Gram negative cell wall
Gram-negative bacterial cell wall is complex. Outer membrane is situated outside the thin peptidoglycan layer. Outer layer is made up of lipopolysaccharide (LPS). LPS are complex molecule consist of three parts. Lipopolysaccharide, also called endo tonics responsible for much of the toxic City e of gram-negative bacterial. LPS elicit strong immune response in animals.
• Lipid A
• Core polysaccharide and O- side chain
Generally the lipid a region contents tu to glucosamine sugar derivative each with three fatty acids and phosphate or pyrophospate attached. Core polysaccharide joined to lipid A.
Microbial evolution Microbial evolution Microbial evolution Microbial evolution Microbial evolution Microbial evolution
अपने दोस्तों के साथ शेयर करे 👇


Recent Comments